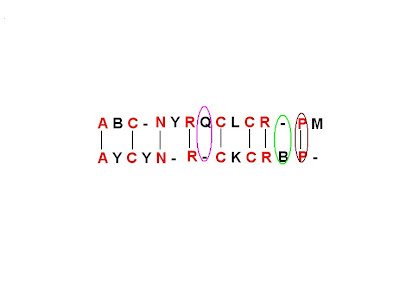Sequence Alignment Techniques are very useful in the field of Bioinformatics. Bioinformatics is the application of computer technology in the field of Biology.
Within the nucleus of our cells, a very fine thread like structure is present known as chromosome. The chief component of chromosome is the DNA( Deoxyribonucleic acid). Without getting into much detail, just know this that the DNA can be represented as a string of letters. Example- ACCTGATCGATCAGTGACGAT, such strings are known as DNA sequences.
Let us align 2 DNA sequences. Seq1- ACTG Seq2- ACTP. Without using any algorithms, we can determine the alignment of these 2 sequence.

Suppose the sequences to be compared are much more complex, say
Seq1- ABCNYRQCLCRPM( Query Sequence )
Seq2- AYCYNRCKCRBP( Subject Sequence )
In such cases, we use algorithms; 2 main algorithms used are Needleman-Wunsch algorithm and Smith-Waterman algorithm. The former is used for Global Alignment while the latter is used for Local alignment. In Global Alignment the whole of the query seqence is compared to the subject sequence for alignment. In Local Alignment, the query sequence is partioned and , and the bits are compared to the subject sequence. Let us solve the above problem using Needleman-Wunsch algorithm.
3 steps are involved-
=> Initiation of the matrix
=> Filling up the matrix
=> Tracing back
=>Initiation of the matrix
Before we initiate the matrix we have to assign the match value, mismatch value and gap value. Let us keep, match=5 mismatch=-3 gap=-4
The initialized matrix is as follows-
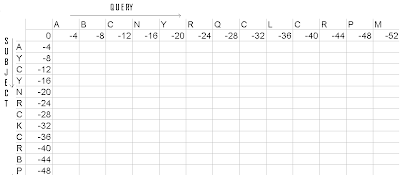
Figure1- Initialized matrix
The query sequence is placed as a row on the top. The subject sequence is placed as a column on the side. 0 is the origin of the matrix. The first row of the matrix is filled with values, as the multiple of the gap value( i.e. -4 is the gap value in this case, so the first row would be filled as -4, -8, -16.... till -52). Do the same for the first column of the matrix.
=> Filling up the matrix
Just consider the labeled cell of the matrix in the figure below. We will fill up this cell upon the basis of fixed protocol. We have to calculate 3 values: (Value of the Diagonal Cell+ Match/Mismatch Value), (Value of the Top Cell+Gap Value), (Value of the Left Cell+ Gap Value). Now we have to select the greatest value, to fill up the cell. In the given example, the 3 values would be, (0+5=5), (-4-4= -8), (-4-4= -8); since 5 is the greatest value, we shall fill up the cell with 5.
Then make another similar matrix. In that fill up the same position, with the type of value you have chosen i.e. - whether the diagonal value is selected, the top value is selected or the left value is selected.
Continue filling both the matrix
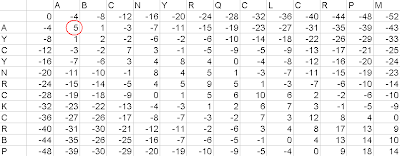
Figure2- Filled up matrix
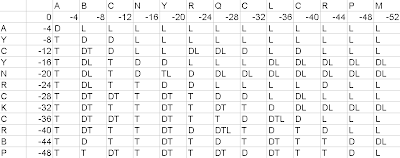
Figure3- Filled up Location matrix
=>Tracing back
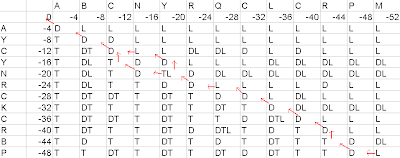
Figure4- Tracing back of the matrix
Based upon the second matrix, we shall trace back. Our start position is the last cell of the last row, and our stopping position should be the first cell of the first row.
If the cell has diagonal value(D), our arrow should move towards the diagonal cell. If the cell has top value(T), our arrow should move towards the top cell. If the cell has left value(L), our arrow should move towards the left cell. If the cell has 2 values(DT or DL or TL), then 2 arrows will be made and they shall move according to the corresponding values. If the cell has 3 values, then 3 arrows will be made and they shall move in all the 3 directions(diagonal, top, left).
After we have obtained the trace back matrix, we shall do the final alignment.
Final Alignment on the basis of first pathway in matrix is:-
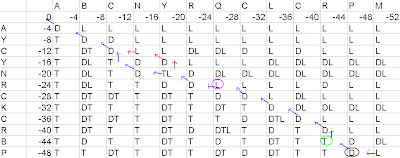
Figure5- First pathway in the matrix
If a cell has diagonal arrow, arising from it, then there will be a match. Example- Just consider the brown labeled cell in the above diagram, since it has a diagonal arrow arising from it, P from the left axis and P from the top axis would be a match.
If the cell has vertical arrow arising from it, then there will be a gap along the query sequence. Example- Just consider the green labeled matrix in the above diagram, since it has a vertical arrow arising from it, there would be a gap along the top axis.
If the cell has a horizontal arrow arising from it, then there will be a gap along the subject sequence. Example- Just consider the pink labeled matrix in the above diagram, since it has a horizontal arrow arising from it, there would be a gap along the left axis.
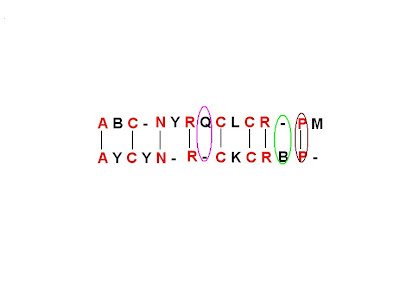
Figure6- Final Result on the basis of the first pathway
Similarly do Final Alignment on the basis of second pathway in matrix:-
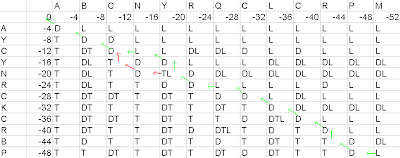
Figure7- Second Result on the basis of the first pathway

Figure8- Final Result on the basis of the second pathway
Both these alignments are correct. I hope you have understood this concept, if not feel free to comment and I shall solve your doubts.